#biomedical




My latest heart.
I know - another heart. I still love making them and, at this time of year, I get a lot of requests. Each is unique - sometimes on purpose, sometimes to cover up an accident - drill scratch, hole in the wrong place etc. The heart itself is vintage - new old stock from the 1970s - a lucite core with a thick copper plate. They are still available but, eventually, I’ll have to find an alternative. I’d love to find similar wood hearts - that could be hinged with a secret internal compartment. If anyone knows where something like that can be sourced - let me know. Dimensions would be 1 ½ inches wide/high and about ¾ of an inch deep so they could be cut and hollowed out.
The next one I’ll be making will be a bit different - a central “window” with a flashing red LED - with batteries that can be replaced. It will have to be a symmetrical design with a feature taking up that much real estate on the heart. I’m starting it tonight and I’ll post a video of it to show the “flash”.

Credit: Purdue University photo/Rebecca McElhoe
By Shardell Joseph
Engineers have developed a handheld paper divide able to detect a strain of coronavirus quickly and accurately. The strain, MERS-CoV, can be detected by the device in very small quantities, and can be read directly from the device making it portable.
As one of the select few given permission by USA health officials to use diagnostics tests for COVID-19, the Purdue University biomedical engineers claimed that this device could be used to detect the COVID-29 strain – the limitations in doing so currently is the lack of funding. According to the team, the process to scale the technology for manufacturing would cost at least a couple of million US dollars.
‘Paper-based devices are already manufactured – pregnancy tests are paper-based,’ said Purdue University Biomedical Engineering Professor, Jacqueline Linnes.
‘Because this device has a more complex shape, a process hasn’t been developed to make it available on a commercial scale. However, many processes in electronics and paper manufacturing could be translated to scaling up this device.’
But so far, Linnes’ team has just been able to produce these devices on a lab scale, which calls for cutting out the paper components by hand.
‘The most difficult aspect of producing this device is definitely the assembly,’ said Purdue University Biomedical Engineering PhD Cadidate, K Byers.

This time-lapse video shows how a paper device developed by Purdue researchers tests a sample in 40 minutes. This sample is positive for MERS-CoV, as indicated by the formation of a second line on the paper strip. Credit: Purdue University video/K Byers
These challenges may easily be overcome with existing manufacturing techniques, the researchers said.
The device format would not need to change in order to detect other diseases. As the device scales up, however, it would also need to be more sensitive to detect a lower concentration of a virus for clinical relevance.
‘This paper device isn’t dependent on a particular virus or sequence. To detect COVID-19, we would just need an assay design specific to that sequence, which could come from a nasal or throat swap sample. Just like with MERS-CoV, a user could load the assay with liquid into the paper platform, fold the device and let it run,’ Linnes said.
When the device folds over, a liquid wash and chemical substances called reagents push the assay up a paper strip to make an easily visible detection line. This automatically completes a multistep process needed for detecting a virus. A user can check the strip within 40 minutes to see if the sample tested positive.
Read the full paper at: https://pubs.acs.org/doi/full/10.1021/acsomega.0c00115?_ga=2.52130409.1579527955.1583773474-378642389.1583773474
Yummy food to keep me going though hell yay




All the stress will end soon… But for some reason I feel like I’ll miss this hectic/messy lifestyle, so I’ll enjoy it so long it lasts.




Only had 2 panic attacks this week… Not bad at all ;D
On a brighter note…. I’ll be graduating (hopefully) very soon, so that makes up for all the uni meltdowns.
I’m pooped
Why am I so unbothered to do any work… While I know I’ll be graduating soon??? HEWLP



Beautiful day in Cambridge, however, my dissertation is due in two weeks so I won’t be able to enjoy the beautiful sunshine this week

⚡Lord Voldermis⚡
A biopsy of a region of skin-that-shall-not-be-named (dermis/hypodermis junction shhh), complete with nerves, vessels, sweat glands and hair follicles.
by @nejiby
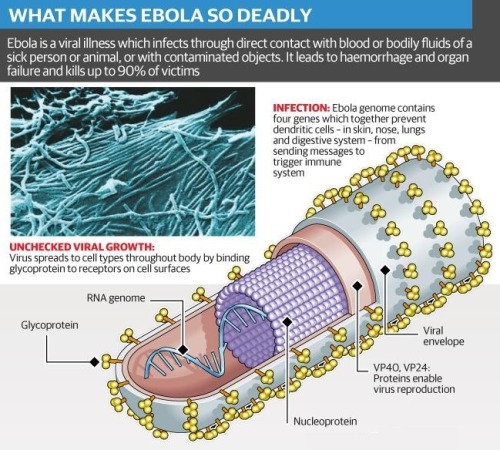
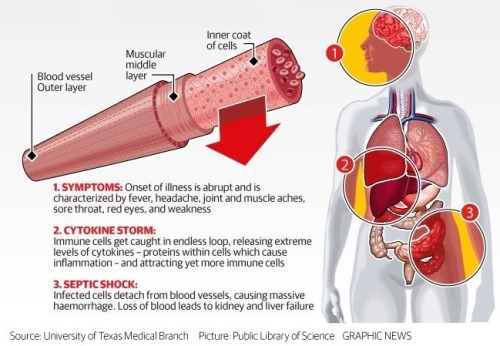
What does Ebola actually do?
By Kelly Servick, 13 August 2014 || Science/AAAS | NewsBehind the unprecedented Ebola outbreak in West Africa lies a species with an incredible power to overtake its host.
Zaire ebolavirus and the family of filoviruses to which it belongs owe their virulence to mechanisms that first disarm the immune response and then dismantle the vascular system.
The virus progresses so quickly that researchers have struggled to tease out the precise sequence of events, particularly in the midst of an outbreak. Much is still unknown, including the role of some of the seven proteins that the virus’s RNA makes by hijacking the machinery of host cells and the type of immune response necessary to defeat the virus before it spreads throughout the body. But researchers can test how the live virus attacks different cells in culture and can observe the disease’s progression in nonhuman primates—a nearly identical model to humans.
Continue reading to find out some of the basic things we understand about how Ebola and humans interact …
IMAGE: The Ebola virus THOMAS W. GEISBERT
Post link
What does Ebola actually do?
By Kelly Servick, 13 August 2014 || Science/AAAS | News
Behind the unprecedented Ebola outbreak in West Africa lies a species with an incredible power to overtake its host.
Zaire ebolavirus and the family of filoviruses to which it belongs owe their virulence to mechanisms that first disarm the immune response and then dismantle the vascular system.
The virus progresses so quickly that researchers have struggled to tease out the precise sequence of events, particularly in the midst of an outbreak. Much is still unknown, including the role of some of the seven proteins that the virus’s RNA makes by hijacking the machinery of host cells and the type of immune response necessary to defeat the virus before it spreads throughout the body. But researchers can test how the live virus attacks different cells in culture and can observe the disease’s progression in nonhuman primates—a nearly identical model to humans.
Continue reading to find out some of the basic things we understand about how Ebola and humans interact …
IMAGE: The Ebola virus THOMAS W. GEISBERT
Post link
STRUCTURE OF CHIKUNGUNYA VIRUS-LIKE PARTICLES
Modeled Using X-ray Crystallography and Electron Cryo-microscopy
Chikungunya, a mosquito-borne virus discovered in 1955 by two scientists in Tanzania, has been a health problem in Africa and southern Asia for decades. It made its way to the Caribbean in late 2013.
Research article: Siyang Sun et al: “Structural analyses at pseudo atomic resolution of Chikungunya virus and antibodies show mechanisms of neutralization” || April 2, 2013 || eLife 2013;2:e00435 [an open access peer reviewed journal]
The Chikungunya virus is carried by mosquitos and can cause a number of diseases in humans including encephalitis, which can be fatal in some cases, and severe arthritis.
Chikungunya virus has a single-stranded RNA genome that codes for four non-structural proteins and five structural proteins. …
A recent mutation in the E1 protein of the virus has allowed it to efficiently reproduce in a different species of mosquitos, leading to a Chikungunya epidemic in Réunion Island in 2005 and the subsequent infection of millions of individuals in Africa and Asia. The virus [has already spread to] the Americas.
[Recently, researchers from Purdue University and several other US institutions, led by Siyang Sun] used two techniques – X-ray crystallography and electron cryo-microscopy – to determine the structure of Chikungunya virus-like particles, and to obtain new insights into the interactions of these particles with four related antibodies.
Electron cryo-microscopy was used to figure out the structure of the particles at near atomic resolution, and X-ray crystallography was used to determine the atomic resolution structures of two of the four Fab [fragment antigen-binding] antibodies that neutralize the Chikungunya virus.
Electron cryo-microscopy was also used to probe the complex formed by the interactions between the virus-like particles and the antibodies.
eLife
Continue readingateLife…
IMAGES
TOP: Structure of the Chikungunya virion ||| BOTTOM: (A) Surface-shaded figure of ectodomain (left) and surface-shaded figure of nucleocapsid (right),colored according to the radial distance from the center of the virus. White triangles indicate one icosahedral asymmetric unit. (B) Cross-section of the virus showing density above 1.5 σ also colored according to the radial distance from the center of the virus. © Resolution of β-strands in the E1 [protein] domain III. [DOI: http://dx.doi.org/10.7554/eLife.00435.003]
SOURCES: Health News : NPR || eLife journal || Wikipedia
Post link
On June 10th there will be a virtual book talk presented by the Penn Libraries’ Biomedical Library and the Kislak Center. “Rediscovering Benjamin Rush, Yellow Fever & the Revolutionary Early Days of Penn Medicine” will feature award- award-winning journalist and New York Times bestselling author Stephen Fried discussing the monumental life of the unsung Founding Father. This talk will be given in conjunction with Fried’s book “RUSH: Revolution, Madness, and Benjamin Rush”, which is available to purchase through our independent bookseller People’s Books & Culture (formerly the Penn Book Center). Registration is required for this free event, please visit the following link for more information: http://ow.ly/DtRS50zSG20
Post link
Sequencing Celebrity Mice: New Study Compares Genetics of 14 Popular Mouse Models
In a new study published on March 9, 2022 in Cell Genomics, researchers at the University of California San Diego School of Medicine present a genome-wide map comparing the genetic makeup of 14 common strains of laboratory mice.
In the century since the C57BL mouse strain was first generated, it has become the most popular laboratory rodent for biomedical research. It functions as a sort of “default” mouse, and its genetic makeup is commonly used as a “neutral” backdrop for genetic modifications that model human diseases. The specific C57BL/6J strain from The Jackson Laboratory is currently the most commonly used inbred mouse, with a closely-related C57BL/10 strain widely used in fields, such as immunology. Many additional sub-strains have since been derived from both.
Given the prevalence of these mouse strains in biology research, a comprehensive understanding of their genetic similarities and differences is valuable to researchers, but until recently, such a resource did not exist.
A team led by Abraham Palmer, PhD, and Jonathan Sebat, PhD, professors at UC San Diego School of Medicine, has now identified 352,631 single nucleotide polymorphisms (SNPs), 109,096 small insertions and deletions (INDELs), 150,344 short tandem repeats (STRs), 3,425 structural variations (SVs) and 2,826 differentially expressed genes (DEGenes) among the different strains. Most of the SNPs were clustered into 28 short segments in the genome, indicating that these genetic differences are likely due to an early introgression of an unrelated mouse, rather than recent independent mutations.
The authors say these results can now be used to guide both forward genetic approaches (wherein scientists identify a phenotypic difference between mice and look for the genetic variation that caused it) and reverse approaches (wherein scientists first identify a genetic difference and then assess whether it produces a different phenotype). Either way, they urge researchers to be aware of the unique genetic profile of their strain of choice.
— Nicole Mlynaryk, Bigelow Science Communication Fellow
Post link